Bioinformatics applications: Difference between revisions
m (Added categories and navbox) |
|||
| Line 141: | Line 141: | ||
Contact: Heng Li <lh3@sanger.ac.uk> | Contact: Heng Li <lh3@sanger.ac.uk> | ||
Usage: bwa <command> [options] | Usage: bwa <command> [options] | ||
[[Category:Guides]] | |||
[[Category:How-Tos]] | |||
{{Navbox Guides}} | |||
Latest revision as of 22:34, 20 September 2023
Strategies to write efficient bioinformatics workflows for the ARC high performance computing (HPC) cluster
One of the challenges to deal with big genomics data set is their long runtimes. The effective and efficient use of the computing resources on the Advanced Research Computing (ARC) cluster can speed up the runtimes.
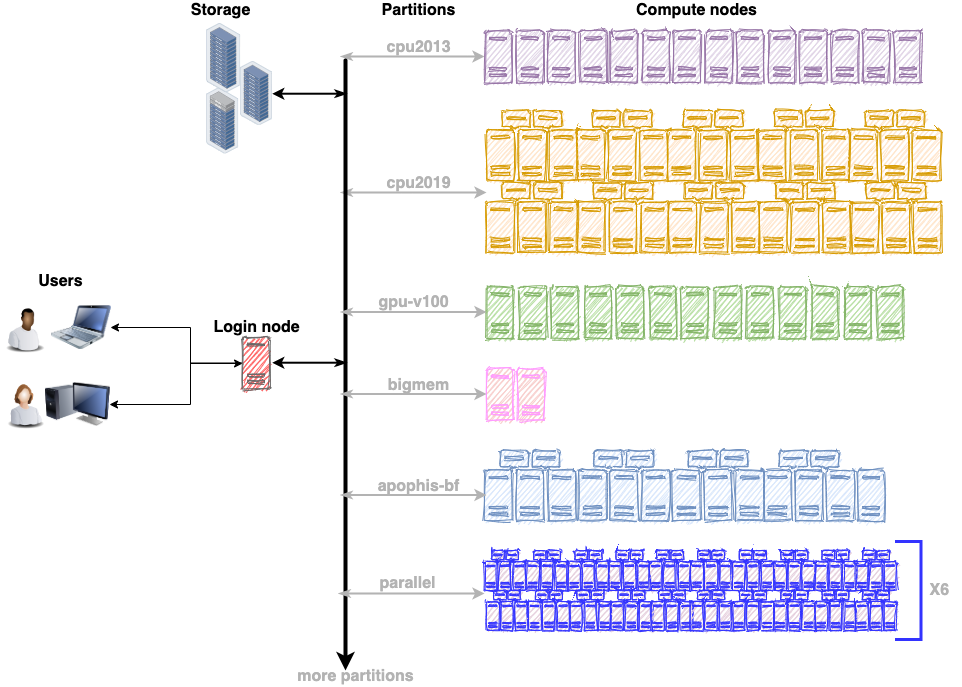
The ARC cluster offers a variety of hardware gears to suit the requirements of any workflow. As a first step, review the compute resources on the ARC cluster (https://rcs.ucalgary.ca/ARC_Cluster_Guide). It will facilitate the process of choosing the appropriate partition depending on the nature of the workflow. For example, choose the gpu-v100 partition for GPU accelerators or the bigmem partition for a memory intensive workflow. Each partition has multiple compute nodes with similar hardware specifications. The illustration below shows that gpu-v100 partition has 13 compute nodes, cpu2019 partition has 40 compute nodes, cpu2013 has 14 compute nodes and so on.
This section will review the SLURM job submission scripts for the bioinformatics applications below.
- FastQC - A high throughput sequence QC analysis tool
- Burrows-Wheeler Aligner (BWA)
- Samtools
FastQC
FastQC assess the quality of your sequencing data. It is available as a module on the ARC cluster. To run the installed version of fastqc, load the biobuilds/2017.11 module as below:
[tannistha.nandi@arc ~]$ module load biobuilds/2017.11
Loading biobuilds/2017.11
Loading requirement: java/1.8.0 biobuilds/conda
[tannistha.nandi@arc ~]$ fastqc --version
FastQC v0.11.5
[tannistha.nandi@arc ~]$ fastqc --help
FastQC - A high throughput sequence QC analysis tool
SYNOPSIS
fastqc seqfile1 seqfile2 .. seqfileN
fastqc [-o output dir] [--(no)extract] [-f fastq|bam|sam] [-c contaminant file] seqfile1..seqfileN
In this example, we will work with the following 35GB of genome sequencing data in fastq file format.
[tannistha.nandi@arc ~]$ ls -l -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 35GB Jan 24. 2018 SN4570284.fq.gz
We will submit the fastqc job on a backfill partition called apophis-bf where each compute node has 40 cores with 185GB of RAM. The job script below requests for 1 core (--cpus-per-task) for 1 hour, along with 300 MB of RAM on the apophis-bf partition. The RAM estimate of 300MB is based on the fastqc user guide that states each thread will be allocated 250MB of memory. On the ARC cluster, the CPUs and cores refer to the same thing. When the job scheduler SLURM will allocate these resources on a compute node, the job will run the first command to load the biobuilds/2017.11 module, which will set the path to fastqc. The next line will launch and run the fastqc application on the data file.
#!/bin/bash #<------------------------Request for Resources-----------------------> #SBATCH –job-name=fastqc-S #SBATCH --mem=300MB #SBATCH –-nodes=1 #SBATCH --ntasks=1 #SBATCH –-cpus-per-task=1 #SBATCH --time= 01:00:00 #SBATCH –-partition=apophis-bf #<------------------------Set environment variables-------------------> module load biobuilds/2017.11 #<------------------------Run python script---------------------------> fastqc -o output --noextract -f fastq SN4570284.fq.gz
Save the above script in a file called ‘fastqc.sh’ and submit it to SLURM using a sbatch command as below.
[tannistha.nandi@arc ~]$ sbatch fastqc.sh Submitted batch job 9658868
The runtime of job id 9658868 is ~46 minutes on a single core with a memory utilization of ~286 MB of RAM.
| Job name | fastqc-S |
|---|---|
| # files | 1 |
| File size | 35GB |
| # cores | 1 |
| CPU efficiency | 99.28% |
| Run time | 46 mins |
| Memory utilized | 286MB |
Strategy to speed up the fastqc runtime
We will use the fastqc runtime flag --threads or -t in short. The flag -t specifies the number of files which can be processed simultaneously. Generally, a single thread will run on 1 core, two threads will run on 2 cores and so on. ‘Fastqc’ can accept multiple input files. One strategy would be to split up the 35GB data file into smaller chunks. Here we have used a python-based tool called ‘fastqsplitter 1.2.0’ (https://pypi.org/project/fastqsplitter/) to split up the fastq file into 10 chunks.
[tannistha.nandi@arc ~]$ salloc –mem=6G -t 01:00:00 -c 10 -N 1 -n 1 -p apophis-bf [tannistha.nandi@cn0513 ~]$ fastqsplitter -t 10 -i SN4570284.fq.gz -o SN4570284_S0.fq.gz -o SN4570284_S1.fq.gz -o SN4570284_S2.fq.gz -o SN4570284_S3.fq.gz -o SN4570284_S4.fq.gz -o SN4570284_S5.fq.gz -o SN4570284_S6.fq.gz -o SN4570284_S7.fq.gz -o SN4570284_S8.fq.gz -o SN4570284_S9.fq.gz [tannistha.nandi@cn0513 ~]$ ls -l -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S0.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S1.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S2.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S3.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S4.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S5.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S6.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S7.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S8.fq.gz -rw-rw-r-- 1 tannistha.nandi tannistha.nandi 3GB May 24 23:15 SN4570284_S9.fq.gz
The core and memory allocation need be adjusted to run fastqc in a multi-threaded mode. The compute resource request will be based on the following factors:
- Number of input files
- Number of cores
- Memory /RAM
- Input file size
There are 10 fastq files, ~3GB each. A total of 10 cores can be requested, one fastq file per core. Accordingly, we need to allocate memory for all 10 cores. A single core fastqc job on a file size of 3GB used ~187 MB of RAM (based on a separate job called 'fastqc-S1'). Thus, we scaled up the memory request for 10 cores to a total of 2000MB of RAM. The multi-threaded job script is saved to a file called ‘fastqc-multicore.sh’.
#!/bin/bash #<------------------------Request for Resources-----------------------> #SBATCH –-jobname=fastqc-M #SBATCH --mem=2000MB #SBATCH –-nodes=1 #SBATCH –-ntasks=1 #SBATCH –-cpus-per-task=10 #SBATCH --time=01:00:00 #SBATCH –-partition=apophis-bf #<------------------------Set environment variables-------------------> module load biobuilds/2017.11 #<------------------------Run python script---------------------------> fastqc --threads 10 -o output_dir --noextract -f fastq SN4570284_S0.fq.gz SN4570284_S1.fq.gz SN4570284_1_S2.fq.gz SN4570284_S3.fq.gz SN4570284_S4.fq.gz SN4570284_S5.fq.gz SN4570284_1_S6.fq.gz SN4570284_S7.fq.gz SN4570284_S8.fq.gz SN4570284_S9.fq.gz
Save the above script as 'fastqc-multicore.sh' and submit it to SLURM using a sbatch command as below:
[tannistha.nandi@arc ~]$ sbatch fastqc-multicore.sh Submitted batch job 9658870
The runtime of multi-threaded fastqc job (job id 9658870) on the same input data size (35G) is ~5 minutes. Thus, the run time of fastqc analysis on the same input size can be reduced significantly using its multi-threaded functionality.
Performance evaluation
It is one of the recommended approaches to reduce the runtime for a given input size. In this example, the multi-threaded feature improved the run time by 10X over a single threaded job. Another point to take note of is that the 4X increased memory utilization of the multi-threaded process. The RAM usage trend is based on the input file size and the way memory allocation is handled by the application.
BWA (Burrows-Wheeler Aligner)
BWA is a popular software for mapping sequencing reads to a reference genome. It is available as a module on the ARC cluster. To run the installed version of bwa, load the biobuilds/2017.11 module as below:
[tannistha.nandi@arc ~]$ module load biobuilds/2017.11 Loading biobuilds/2017.11 Loading requirement: java/1.8.0 biobuilds/conda [tannistha.nandi@arc ~]$ bwa Program: bwa (alignment via Burrows-Wheeler transformation) Version: 0.7.17-r1188 Contact: Heng Li <lh3@sanger.ac.uk> Usage: bwa <command> [options]