Running jobs
This page is intended for users who are looking for guidance on working effectively with the SLURM job scheduler on the compute clusters that are operated by Research Computing Services (RCS). The content on this page assumes that you already familiar with the basic concepts of job scheduling and job scripts. If you have not worked on a large shared computer cluster before, please read What is a scheduler? first. To familiarize yourself with how we use computer architecture terminology in relation to SLURM, you may also want to read about the Types of Computational Resources, as well as the cluster guide for the cluster that you are working on. Users familiar with other job schedulers including PBS/Torque, SGE, LSF, or LoadLeveler, may find this table of corresponding commands useful. Finally, if you have never done any kind of parallel computation before, you may find some of the content on Parallel Models useful as an introduction.
Basic HPC Workflow
The most basic HPC computation workflow (excluding software setup and data transfer) involves 5 steps.
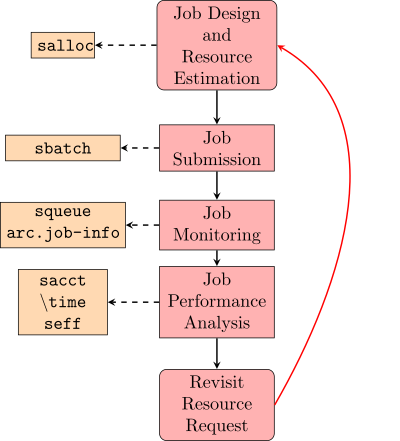
This process begins with the creation of a working preliminary job script that includes an approximate resource request and all of the commands required to complete a computational task of interest. This job script is used as the basis of a job submission to SLURM. The submitted job is then monitored for issues and analyzed for resource utilization. Depending on how well the job utilized resources, modifications to the job script can be made to improve computational efficiency and the accuracy of the resource request. In this way, the job script can be iteratively improved until a reliable job script is developed. As your computational goals and methods change, your job scripts will need to evolve to reflect these changes and this workflow can be used again to make appropriate modifications.
Job Design and Resource Estimation
The goal of this first step is the development of a slurm job script for the calculations that you wish to run that reflects real resource usage in a job typical of your work. To review some example jobs scripts for different models of parallel computation, please consult the page Sample Job Scripts. We will look at the specific example of using the NumPy library in python to multiply two matrices that are read in from two files. We assume that the files have been transferred to a subdirectory of the users home directory ~/projects/matmul and that a custom python installation has been setup under ~/anaconda3.
$ ls -lh ~/project/matmul
total 4.7G
-rw-rw-r-- 1 username username 2.4G Feb 3 10:13 A.csv
-rw-rw-r-- 1 username username 2.4G Feb 3 10:16 B.csv
$ ls ~/anaconda3/bin/python
/home/username/anaconda3/bin/python
The computational plan for this project is to load the data from files and then use the multithreaded version of the NumPy dot function to perform the matrix multiplication and write it back out to a file. To test out our proposed steps and software environment, we will request a small Interactive Job. This is done with the salloc command. The salloc command accepts all of the standard resource request parameters for SLURM. (The key system resources that can be requested can be reviewed on the Types of Computational Resources page) In our case, we will need to specify a partition, a number of cores, a memory allocation, and a small time interval in which we will do our testing work (on the order of 1-4 hours). We begin by checking which partitions are currently not busy using the arc.nodes command
$ arc.nodes
Partitions: 16 (apophis, apophis-bf, bigmem, cpu2013, cpu2017, cpu2019, gpu-v100, lattice, parallel, pawson, pawson-bf, razi, razi-bf, single, theia, theia-bf)
====================================================================================
| Total Allocated Down Drained Draining Idle Maint Mixed
------------------------------------------------------------------------------------
apophis | 21 1 1 0 0 7 1 11
apophis-bf | 21 1 1 0 0 7 1 11
bigmem | 2 0 0 0 0 1 0 1
cpu2013 | 14 1 0 0 0 0 0 13
cpu2017 | 16 0 0 0 0 0 16 0
cpu2019 | 40 3 1 2 1 0 4 29
gpu-v100 | 13 2 0 1 0 5 0 5
lattice | 279 34 3 72 0 47 0 123
parallel | 576 244 26 64 1 20 3 218
pawson | 13 1 1 2 0 9 0 0
pawson-bf | 13 1 1 2 0 9 0 0
razi | 41 26 4 1 0 8 0 2
razi-bf | 41 26 4 1 0 8 0 2
single | 168 0 14 31 0 43 0 80
theia | 20 0 0 0 1 0 0 19
theia-bf | 20 0 0 0 1 0 0 19
------------------------------------------------------------------------------------
logical total | 1298 340 56 176 4 164 25 533
|
physical total | 1203 312 50 173 3 140 24 501
The ARC Cluster Guide tells us that the single partition is an old partition with 12GB of Memory (our data set is only 5GB and matrix multiplication is usually pretty memory efficient) and 8 cores so it is probably a good fit. Based on the arc.nodes output, we can choose the single partition (which at the time that this ran had 43 idle nodes) for our test. The preliminary resource request that we will use for the interactive job is
partition=single time=2:0:0 nodes=1 ntasks=1 cpus-per-task=8 mem=0
We are requesting 2 hours and 0 minutes and 0 seconds, a whole single node (i.e. all 8 cores), and mem=0 means that all memory on the node is being requested. Please note that mem=0 can only used in a memory request when you are already requesting all of the cpus on a node. On the single partition, 8 cpus is the full number of cpus per node. Please check this for the partition that you plan on running on using arc.hardware before using it. From the login node, we will request this job using salloc
[userrname@arc ~]$ salloc --partition=single --time=2:0:0 --nodes=1 --ntasks=1 --cpus-per-task=8 --mem=8gb
salloc: Pending job allocation 8430460
salloc: job 8430460 queued and waiting for resources
salloc: job 8430460 has been allocated resources
salloc: Granted job allocation 8430460
salloc: Waiting for resource configuration
salloc: Nodes cn056 are ready for job
[username@cn056 ~]$ export PATH=~/anaconda3/bin:$PATH
[username@cn056 ~]$ which python
~/anaconda3/bin/python
[username@cn056 ~]$ export OMP_NUM_THREADS=4
[username@cn056 ~]$ export OPENBLAS_NUM_THREADS=4
[username@cn056 ~]$ export MKL_NUM_THREADS=4
[username@cn056 ~]$ export VECLIB_MAXIMUM_THREADS=4
[username@cn056 ~]$ export NUMEXPR_NUM_THREADS=4
[username@cn056 ~]$ python
Python 3.7.4 (default, Aug 13 2019, 20:35:49)
[GCC 7.3.0] :: Anaconda, Inc. on linux
Type "help", "copyright", "credits" or "license" for more information.
>>>
The steps that were taken at the command line included
- submitting an interactive job request and waiting for the scheduler to allocate the node (cn056) and move the terminal to that node
- setting up our software environment using Environment Variables
- starting a python interpreter
At this point, we are ready to test our simple python code at the python interpreter command line.
>>> import numpy as np
>>> A=np.loadtxt("/home/username/project/matmul/A.csv",delimiter=",")
>>> B=np.loadtxt("/home/username/project/matmul/B.csv",delimiter=",")
>>> C=np.dot(A,B)
>>> np.savetxt("/home/username/project/matmul/C.csv",C,delimiter=",")
>>> quit()
[username@cn056 ~]$ exit
exit
salloc: Relinquishing job allocation 8430460
In a typical testing/debugging/prototyping session like this, some problems would be identified and fixed along the way. Here we have only shown successful steps of a simple job. However, normally this would be an opportunity to identify syntax errors, data type mismatch, out-of-memory exceptions, and other issues in an interactive environment. The data used for a test should be a manageable size. In the sample data, I am working with randomly generated 10000x10000 matrices and discussing it as though it was the actual job that would run. However, often times, the real problem is too large for an interactive job and it is better to test the commands in an interactive mode using a small data subset. For example, randomly generated 10000 x 10000 matrices might be a good starting point for planning a real job that was going to take the product of 1 million x1 million matrices.
While this interactive job was running, I used a separate terminal on ARC to check the resource utilization of the interactive job by running the command arc.job-info 8430460, and found that the CPU utilization hovered around 50% and memory utilization around 30%. We will come back to this point in later steps of the workflow but this would be a warning sign that the resource request is a bit high, especially in CPUs. It is worth noting that if you have run your code before on a different computer, you can base your initial slurm script on that past experience or the resources that are recommended in the software documentation instead of testing it out explicitly with salloc. However, as the the hardware and software installation are likely different, you may run into new challenges that need to be resolved. In the next step, we will write a slurm script that includes all of the commands we ran manually in the above example.
Job Submission
In order to use the full resources of a computing cluster, it is important to do your large-scale work with Batch Jobs. This amounts to replacing sequentially typed commands (or successively run cells in a notebook) with a shell script that automates the entire process and using the sbatch command to submit the shell script as a job to the job scheduler. Please note, you must not run this script directly on the login node. It must be submitted to the job scheduler.
Simply putting all of our commands in a single shell script file (with proper #SBATCH directives for the resource request), yields the following unsatisfactory result:
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=4
#SBATCH --mem=8gb
#SBATCH --time=2:0:0
export PATH=~/anaconda3/bin:$PATH
echo $(which python)
export OMP_NUM_THREADS=4
export OPENBLAS_NUM_THREADS=4
export MKL_NUM_THREADS=4
export VECLIB_MAXIMUM_THREADS=4
export NUMEXPR_NUM_THREADS=4
python matmul_test.py
where matmul_test.py:
import numpy as np
A=np.loadtxt("/home/username/project/matmul/A.csv",delimiter=",")
B=np.loadtxt("/home/username/project/matmul/B.csv",delimiter=",")
C=np.dot(A,B)
np.savetxt("/home/username/project/matmul/C.csv",C,delimiter=",")
There are a number of things to improve here when moving to a batch job. We shoul consider generalizing our script a bit to improve reusability between jobs, instead of hard coding paths. This yields:
matmul_test02032021.slurm:
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=4
#SBATCH --mem=10000M
#SBATCH --time=2:0:0
export PATH=~/anaconda3/bin:$PATH
echo $(which python)
export OMP_NUM_THREADS=4
export OPENBLAS_NUM_THREADS=4
export MKL_NUM_THREADS=4
export VECLIB_MAXIMUM_THREADS=4
export NUMEXPR_NUM_THREADS=4
AMAT="/home/username/project/matmul/A.csv"
BMAT="/home/username/project/matmul/B.csv"
OUT="/home/username/project/matmul/C.csv"
python matmul_test.py $AMAT $BMAT $OUT
where matmul_test.py:
import numpy as np
import sys
#parse arguments
if len(sys.argv)<3:
raise Exception
lmatrixpath=sys.argv[1]
rmatrixpath=sys.argv[2]
outpath=sys.argv[3]
A=np.loadtxt(lmatrixpath,delimiter=",")
B=np.loadtxt(rmatrixpath,delimiter=",")
C=np.dot(A,B)
np.savetxt(outpath,C,delimiter=",")
We have generalized the way that data sources and output locations are specified in the job. In the future, this will help us reuse this script without proliferating python scripts. Similar considerations of reusability apply to any language and any job script. In general, due to the complexity of using them, HPC systems are not ideal for running a couple small calculations. They are at their best when you have to solve many variations on a problem at a large scale. As such, it is valuable to always be thinking about how to make your workflows simpler and easier to modify.
Now that we have job script, we can simply submit the script to sbatch:
[username@arc matmul]$ sbatch matmul_test02032021.slurm
Submitted batch job 8436364
The number reported after the sbatch job submission is the JobID and will be used in all further analyses and monitoring. With this number, we can begin the process of checking on how our job progresses through the scheduling system.
Job Monitoring
The third step in this work flow is checking the progress of the job. We will emphasize three tools:
--mail-user,--mail-type- sbatch options for sending email about job progress
- no performance information
squeue- slurm tool for monitoring job start, running, and end
- no performance information
arc.job-info- RCS tool for monitoring job performance once it is running
- provides detailed snapshot of key performance information
The motives for using the different monitoring tools are very different but they all help you track the progress of your job from start to finish. The details of these tools applied to the above example are discussed in the article Job Monitoring. If a job is determined to be incapable of running or have a serious error in it, you can cancel it at any time using the command scancel JobID, for example scancel 8442969
Further analysis of aggregate and maximum resource utilization is needed to determine changes that need to be made to the job script and the resource request. This is the focus of the Job Performance Analysis step that is the subject of the next section.
Job Performance Analysis
There are a tremendous number of performance analysis tools, from complex instruction-analysis tools like gprof, valgrind, and nvprof to extremely simple tools that only provide average utilization. The minimal tools that are required for characterizing a job are
\timefor estimating CPU utilization on a single nodesacct -j JobID -o JobIDMaxRSS,Elapsedfor analyzing maximum memory utilization and wall timeseff JobIDfor analyzing general resource utilization
When utilizing these tools, it is important to consider the different sources of variability in job performance. Specifically, it is important to perform a detailed analysis of
- Variability Among Datasets
- Variability Across Hardware
- Request Parameter Estimation from Samples Sets - Scaling Analysis
and how these can be addressed through thoughtful job design. This is the subject of the article Job Performance Analysis and Resource Planning.
Revisiting the Resource Request
Having completed a careful analysis of performance for our problem and the hardware that we plan to run on, we are now in a position to revise the resource request and submit more jobs. If you have 10000 such jobs then it is important to think about how busy the resources that you are planning to use are likely to be. It is also important to think about the fact that every job has some finite scheduling overhead (time to setup the environment to run in, time to figure out where to run to optimally fit jobs, etc). In order to make this scheduling overhead worthwhile, it is generally important to have jobs run for more than 10 minutes. So, we may want to pack batches of 10-20 matrix pairs into one job just to make it a sensible amount of work. This also will have some benefit in making job duration more predictable when there is variability due to data sets. New hardware also tends to be busier than old hardware and cpu2019 will often have 24 hour wait times when the cluster is busy. However, for jobs that can stay under 5 hours, the backfill partitions are often not busy and so the best approach might be to bundle 20 tasks together for a duration of about 100 minutes and then have the job request be
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=4
#SBATCH --mem=5000M
#SBATCH --time=1:40:0
#SBATCH --array=1-500
export PATH=~/anaconda3/bin:$PATH
echo $(which python)
export OMP_NUM_THREADS=4
export OPENBLAS_NUM_THREADS=4
export MKL_NUM_THREADS=4
export VECLIB_MAXIMUM_THREADS=4
export NUMEXPR_NUM_THREADS=4
export TOP_DIR="/home/username/project/matmul/pairs"$SLURM_ARRAY_TASK_ID
DIR_LIST=$(find $TOP_DIR -type d -mindepth 1)
for DIR in $FLIST
do
python matmul_test.py $DIR/A.csv $DIR/B.csv $DIR/C.csv;
done
Here the directory TOP_DIR will be one of 500 directories, each named pairsN where N is an integer from 1 to 500, with each of these holding 20 subdirectories with a pair of A and B matrix files in each. The script will iterate through the 20 subdirectories and run the python script once for each pair of matrices. This is one approach to organizing work into bundles. You could then submit a job array The choice depends on a lot of assumptions that we have made, but it is a fairly good plan. The result would be 500 jobs, each 2 hours long, running mostly on the backfill partitions on 1 core a piece. If there is usually at least 1 node free, that will be about 38 jobs running at the same time for a total of roughly 13 sequences that are 2 hours long, or 26 hours. In this way, all of work could be done in a day. Typically, we would iteratively improve the script until it was something that could scale up. This is why the workflow is a loop.
Tips and tricks
Attaching to a running job
It is possible to connect to the node running a job and execute new processes there. You might want to do this for troubleshooting or to monitor the progress of a job.
Suppose you want to run the utility nvidia-smi to monitor GPU usage on a node where you have a job running.
The following command runs watch on the node assigned to the given job, which in turn runs nvidia-smi every 30 seconds,
displaying the output on your terminal.
$ srun --jobid 123456 --pty watch -n 30 nvidia-smi
It is possible to launch multiple monitoring commands using tmux.
The following command launches htop and nvidia-smi in separate panes to monitor the activity on a node assigned to the given job.
$ srun --jobid 123456 --pty tmux new-session -d 'htop -u $USER' \; split-window -h 'watch nvidia-smi' \; attach
Processes launched with srun share the resources with the job specified. You should therefore be careful not to launch processes that would use a significant portion of the resources allocated for the job. Using too much memory, for example, might result in the job being killed; using too many CPU cycles will slow down the job.
Noteː The srun commands shown above work only to monitor a job submitted with sbatch. To monitor an interactive job, create multiple panes with tmux and start each process in its own pane.
It is possible to attach second terminal to a running interactive job with the
$ srun -s --pty --jobid=12345678 /bin/bash
The -s option specifies that the resources can be oversubscribed. It is needed as all the resources have already be given to the first shell. So, to run the second shell we either have to wait until the first shell is over, or we have to oversubscribe.
Checkpointing
Checkpointing refers to the technique of storing a running program's execution state so that it can restart in the exact same situation later on or on another machine. This is applicable to serial or parallel/distributed computing on CPUs and GPUs. Some HPC applications include checkpointing as a feature, in many other cases, such as custom scripts and code, the developer is expected to implement their own checkpointing solution that save the state of the computation and resume from it in a separate job.
Checkpointing is useful for various reasons, for example:
- Many compute partitions enforce strict wall-time limits. On ARC, GPU partitions have a maximum runtime of 24 hours, while CPU partitions are limited to 7 days. Jobs are automatically terminated once these limits are reached. By using checkpointing, long-running applications can be executed across multiple job submissions without loss of progress.
- Checkpointing allows applications to recover from failures by restarting from the most recent saved state rather than from the beginning. Job failures may occur due to:
- Application-level errors, such as exceeding memory limits or encountering runtime exceptions.
- A job might also fail dues to System-level issues, including hardware failures or problems affecting the compute node.
Further reading
- Slurm Manual - https://slurm.schedmd.com/
- Additional tutorials.
- sbatch command options.
- A "Rosetta stone" mapping commands and directives from PBS/Torque, SGE, LSF, and LoadLeveler, to SLURM.
- http://www.ceci-hpc.be/slurm_tutorial.html - Text tutorial from CÉCI, Belgium
- http://www.brightcomputing.com/blog/bid/174099/slurm-101-basic-slurm-usage-for-linux-clusters - Minimal text tutorial from Bright Computing