ARC Cluster Guide: Difference between revisions
m added link for backing up ARC data to OneDrive |
|||
| Line 158: | Line 158: | ||
Please contact us at support@hpc.ucalgary.ca if you want more information about this option. | Please contact us at support@hpc.ucalgary.ca if you want more information about this option. | ||
You can also back up data to your UofC OneDrive for business allocation see: https://rcs.ucalgary.ca/How_to_transfer_data#rclone:_rsync_for_cloud_storage This allocation starts at 5TB. Contact the support center for questions regarding OneDrive for Business. | |||
}} | }} | ||
Revision as of 14:29, 9 July 2021
|
Cybersecurity awareness at the U of C Please note that there are typically about 950 phishing attempts targeting University of Calgary accounts each month. This is just a reminder to be careful about computer security issues, both at home and at the University. Please visit https://it.ucalgary.ca/it-security for more information, tips on secure computing, and how to report suspected security problems.
|
|
Need Help or have other ARC Related Questions? For all general RCS related issues, questions, or comments, please contact us at support@hpc.ucalgary.ca.
|
This guide gives an overview of the Advanced Research Computing (ARC) cluster at the University of Calgary and is intended to be read by new account holders getting started on ARC. This guide covers topics such as the hardware and performance characteristics, available software, usage policies and how to log in and run jobs.
Introduction
The ARC compute cluster can be used for running large numbers (hundreds) of concurrent serial (one core) jobs, OpenMP or other thread-based jobs, shared-memory parallel code using up to 40 or 80 threads per job (depending on the partition), distributed-memory (MPI-based) parallel code using up to hundreds of cores, or jobs that take advantage of Graphics Processing Units (GPUs). Almost all work on ARC is done through a command line interface. This computational resource is available for research projects based at the University of Calgary and is meant to supplement the resources available to researchers through Compute Canada.
Historically, ARC is primarily comprised of older, disparate Linux-based clusters that were formerly offered to researchers from across Canada such as Breezy, Lattice, and Parallel. In addition, a large-memory compute node (Bigbyte) was salvaged from the now-retired local Storm cluster. In January 2019, a major addition to ARC with modern hardware was purchased. In 2020, compute clusters from CHGI have been migrated into ARC.
How to Get Started
If you have a project you think would be appropriate for ARC, please write to support@hpc.ucalgary.ca and mention the intended research and software you plan to use. You must have a University of Calgary IT account in order to use ARC.
- For users that do not have a University of IT account or email address, please register for one at https://itregport.ucalgary.ca/.
- For users external to the University, such as for users collaborating on a research project at the University of Calgary, please contact us and mention the project leader you are collaborating with.
Once your access to ARC has been granted, you will be able to immediately make use of the cluster using your University of Calgary IT account by following the usage guide outlined below.
Hardware
Since the ARC cluster is a conglomeration of many different compute clusters, the hardware within ARC can vary widely in terms of performance and capabilities. To mitigate any compatibility issues with different hardware, we combine similar hardware into their own Slurm partition to ensure your workload runs as consistently as possible within one partition. Please carefully review the hardware specs for each of the partitions below to avoid any surprises.
Partition Hardware Specs
When submitting jobs to ARC, you may specify a partition that your job will run on. Please choose a partition that is most appropriate for your work.
A few things to keep in mind when choosing a partition:
- Specific workloads requiring special Intel Instruction Set Extensions may only work on newer Intel CPUs.
- If working with multi-node parallel processing, ensure your software and libraries support the partition's interconnect networking.
- While older partitions may be slower, they may be less busy and have little to no wait times.
If you are unsure which partition to use or need assistance on selecting an appropriate partition, please see the Selecting a Partition Section below.
| Partition | Description | Nodes | CPU Cores, Model, and Year | Memory | GPU | Network |
|---|---|---|---|---|---|---|
| - | ARC Login Node | 1 | 16 cores, 2x Intel(R) Xeon(R) CPU E5620 @ 2.40GHz (2010) | 48 GB | N/A | 40 Gbit/s InfiniBand |
| gpu-v100 | GPU Parition | 13 | 80 cores, 2x Intel(R) Xeon(R) Gold 6148 CPU @ 2.40GHz (2019) | 754 GB | 2x Tesla V100-PCIE-16GB | 100 Gbit/s Omni-Path |
| cpu2019 | General Purpose Compute | 14 | 40 cores, 2x Intel(R) Xeon(R) Gold 6148 CPU @ 2.40GHz (2019) | 190 GB | N/A | 100 Gbit/s Omni-Path |
| apophis | General Purpose Compute | 21 | 40 cores, 2x Intel(R) Xeon(R) Gold 6148 CPU @ 2.40GHz (2019) | 190 GB | N/A | 100 Gbit/s Omni-Path |
| razi | General Purpose Compute | 41 | 40 cores, 2x Intel(R) Xeon(R) Gold 6148 CPU @ 2.40GHz (2019) | 190 GB | N/A | 100 Gbit/s Omni-Path |
| bigmem | Big Memory Nodes | 2 | 80 cores, 4x Intel(R) Xeon(R) Gold 6148 CPU @ 2.40GHz (2019) | 3022 GB | N/A | 100 Gbit/s Omni-Path |
| pawson | General Purpose Compute | 13 | 40 cores, 2x Intel(R) Xeon(R) Gold 6248 CPU @ 2.50GHz (2019) | 190 GB | N/A | 100 Gbit/s Omni-Path |
| cpu2017 | General Purpose Compute | 14 | 56 cores, 2x Intel(R) Xeon(R) CPU E5-2690 v4 @ 2.60GHz (2016) | 256 GB | N/A | 40 Gbit/s InfiniBand |
| theia | Former Theia cluster | 20 | 56 cores, 2x Intel(R) Xeon(R) CPU E5-2690 v4 @ 2.60GHz (2012) | 188 GB | N/A | 40 Gbit/s InfiniBand |
| cpu2013 | Former hyperion cluster | 12 | 32 cores, 2x Intel(R) Xeon(R) CPU E5-2670 0 @ 2.60GHz (2012) | 126 GB | N/A | 40 Gbit/s InfiniBand |
| lattice | Former Lattice cluster | 307 | 8 cores, 2x Intel(R) Xeon(R) CPU L5520 @ 2.27GHz (2011) | 12 GB | N/A | 40 Gbit/s InfiniBand |
| single | Former Lattice cluster | 168 | 8 cores, 2x Intel(R) Xeon(R) CPU L5520 @ 2.27GHz (2011) | 12 GB | N/A | 40 Gbit/s InfiniBand |
| parallel | Former Parallel Cluster | 576 | 12 cores, 2x Intel(R) Xeon(R) CPU E5649 @ 2.53GHz (2011) | 24 GB | N/A | 40 Gbit/s InfiniBand |
ARC Cluster Storage
Usage of ARC cluster storage is outlined by our ARC Storage Terms of Use page.
|
No Backup Policy! You are responsible for your own backups. Many researchers will have accounts with Compute Canada and may choose to back up their data there (the Project file system accessible through the Cedar cluster would often be used).
Please contact us at support@hpc.ucalgary.ca if you want more information about this option. You can also back up data to your UofC OneDrive for business allocation see: https://rcs.ucalgary.ca/How_to_transfer_data#rclone:_rsync_for_cloud_storage This allocation starts at 5TB. Contact the support center for questions regarding OneDrive for Business. |
The ARC cluster has around 2 petabyte of shared disk storage available across the entire cluster as well as temporary storage local to each of the compute nodes. Please refer to the individual sections below on the capacity limitations and usage policies.
Use the arc.quota command on ARC to determine the available space on your various volumes and home directory.
| Partition | Description | Capacity |
|---|---|---|
/home
|
User home directories | 500 GB (per user) |
/work
|
Research project storage | Up to 100's of TB |
/scratch
|
Scratch space for temporary files | Up to 30 TB |
/tmp
|
Temporary space local to the compute cluster | Dependent on nodes, use df -h.
|
/dev/shm
|
Small temporary in-memory disk space local to the compute cluster | Dependent on nodes, use df -h.
|
/home: Home file system
Each user has a directory under /home and is the default working directory when logging in to ARC. Each home directory has a per-user quota of 500 GB. This limit is fixed and cannot be increased. Researchers requiring additional storage exceeding what is available on their home directory may use /work and /scratch.
Note on file sharing: Due to security concerns, permissions set using chmod on your home directory to allow other users to read/write to your home directory be automatically reverted by an automated system process unless an explicit exception is made. If you need to share files with other researchers on the ARC cluster, please write to support@hpc.ucalgary.ca to ask for such an exception.
/scratch: Scratch file system for large job-oriented storage
Associated with each job, under the /scratch directory, a subdirectory is created that can be referenced in job scripts as /scratch/${SLURM_JOB_ID}. You can use that directory for temporary files needed during the course of a job. Up to 30 TB of storage may be used, per user (total for all your jobs) in the /scratch file system.
Data in /scratch associated with a given job will be deleted automatically, without exception, five days after the job finishes.
/work: Work file system for larger projects
If you need more space than provided in /home and the /scratch job-oriented space is not appropriate for you case, please write to support@hpc.ucalgary.ca with an explanation, including an indication of how much storage you expect to need and for how long. If approved, you will then be assigned a directory under /work with an appropriately large quota.
/tmp, /dev/shm: Temporary files
You may use /tmp for temporary files generated by your job. The /tmp is stored on a disk local to the compute node and is not shared across the cluster. The files stored here may be removed immediately after your job terminates.
/dev/shm is similar to /tmp but the storage is backed by virtual memory for higher IOPS. This is ideal for workloads that require many small read/writes to share data between processes or as a fast cache. The files stored here may be removed immediately after your job terminates.
Using ARC
Logging in
To log in to ARC, connect using SSH to arc.ucalgary.ca on port 22. Connections to ARC are accepted only from the University of Calgary network (on campus) or through the University of Calgary General VPN (off campus).
See Connecting to RCS HPC Systems for more information.
How to interact with ARC
ARC cluster is a collection of several compute nodes connected by a high-speed network. On ARC, computations get submitted as jobs. Once submitted, the jobs are then assigned to compute nodes by the job scheduler as resources become available.
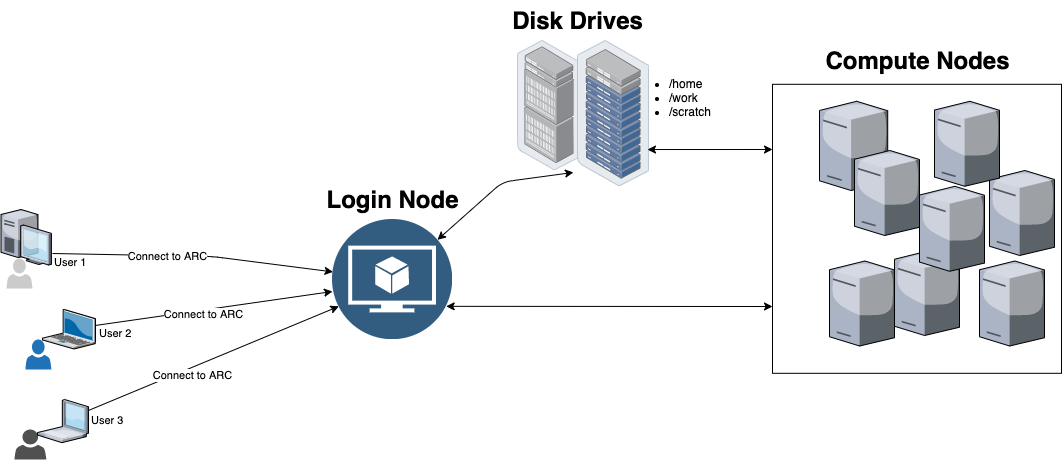
You can access ARC with your UCalgary IT user credentials. Once connected, you will get placed in the ARC login node, for basic tasks such as job submission, monitor job status, manage files, edit text, etc. It is a shared resource where multiple users get connected at the same time. Thus, any intensive tasks is not allowed on the login node as it may block other potential users to connect/submit their computations.
[tannistha.nandi@arc ~]$
The job scheduling system on ARC is called SLURM. On ARC, there are two SLURM commands that can allocate resources to a job under appropriate conditions: ‘salloc’ and ‘sbatch’. They both accept the same set of command line options with respect to resource allocation.
‘salloc’ is to launch an interactive session, typically for tasks under 5 hours. Once an interactive job session is created, you can do things like explore research datasets, start R or python sessions to test your code, compile software applications etc.
a. Example 1: The following command requests for 1 cpu on 1 node for 1 task along with 1 GB of RAM for an hour.
[tannistha.nandi@arc ~]$ salloc --mem=1G -c 1 -N 1 -n 1 -t 01:00:00
salloc: Granted job allocation 6758015
salloc: Waiting for resource configuration
salloc: Nodes fc4 are ready for job
[tannistha.nandi@fc4 ~]$
b. Example 2: The following command requests for 1 GPU to be used from 1 node belonging to the gpu-v100 partition along with 1 GB of RAM for 1 hour. Generic resource scheduling (--gres) is used to request for GPU resources.
[tannistha.nandi@arc ~]$ salloc --mem=1G -t 01:00:00 -p gpu-v100 --gres=gpu:1
salloc: Granted job allocation 6760460
salloc: Waiting for resource configuration
salloc: Nodes fg3 are ready for job
[tannistha.nandi@fg3 ~]$
Once you finish the work, type 'exit' at the command prompt to end the interactive session,
[tannistha.nandi@fg3 ~]$ exit
[tannistha.nandi@fg3 ~]$ salloc: Relinquishing job allocation 6760460
It is to ensure that the allocated resources are released from your job and now available to other users.
‘sbatch’ is to submit computations as jobs to run on the cluster. You can submit a job-script.slurm via 'sbatch' for execution.
[tannistha.nandi@arc ~]$ sbatch job-script.slurm
When resources become available, they get allocated to this task. Batch jobs are suited for tasks that run for long periods of time without any user supervision. When the job-script terminates, the allocation is released. Please review the section on how to prepare job scripts for more information.
Prepare job scripts
Job scripts are text files saved with an extension '.slurm', for example, 'job-script.slurm'. A job script looks something like this:
#!/bin/bash ####### Reserve computing resources ############# #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --cpus-per-task=1 #SBATCH --time=01:00:00 #SBATCH --mem=1G #SBATCH --partition=cpu2019
####### Set environment variables ############### module load python/anaconda3-2018.12
####### Run your script ######################### python myscript.py
The first line contains the text "#!/bin/bash" to interpret it as a bash script.
It is followed by lines that start with a '#SBATCH' to communicate with 'SLURM'. You may add as many #SBATCH directives as needed to reserve computing resources for your task. The above example requests for one cpu on a single node for 1 task along with 1GB RAM for an hour on cpu2019 partition.
Next, you have to set up environment variables either by loading the modules centrally installed on ARC or export path to the software in your home directory. The above example loads an available python module.
Finally, include the Linux command to execute the local script.
Note that failing to specify part of a resource allocation request (most notably time and memory) will result in bad resource requests as the defaults are not appropriate to most cases. Please refer to the section 'Running non-interactive jobs' for more examples.
Software
All ARC nodes run the latest version of CentOS 7 with the same set of base software packages. To maintain the stability and consistency of all nodes, any additional dependencies that your software requires must be installed under your account. For your convenience, we have packaged commonly used software packages and dependencies as modules available under /global/software. If your software package is not available as a module, you may also try Anaconda which allows users to manage and install custom packages in an isolated environment.
For a list of available packages that have been made available, please see ARC Software pages.
Please contact us at support@hpc.ucalgary.ca if you need additional software installed.
Modules
The setup of the environment for using some of the installed software is through the module command. An overview of modules on WestGrid (external link) is largely applicable to ARC.
Software packages bundled as a module will be available under /global/software and can be listed with the module avail command.
$ module avail
To enable Python, load the Python module by running:
$ module load python/anaconda-3.6-5.1.0
To unload the Python module, run:
$ module remove python/anaconda-3.6-5.1.0
To see currently loaded modules, run:
$ module list
By default, no modules are loaded on ARC. If you wish to use a specific module, such as the Intel compilers or the Open MPI parallel programming packages, you must load the appropriate module.
Storage
Please review the Storage section above for important policies and advice regarding file storage and file sharing.
Interactive Jobs
The ARC login node may be used for such tasks as editing files, compiling programs and running short tests while developing programs. We suggest CPU intensive workloads on the login node be restricted to under 15 minutes as per our cluster guidelines. For interactive workloads exceeding 15 minutes, use the salloc command to allocate an interactive session on a compute node.
The default salloc allocation is 1 CPU and 1 GB of memory. Adjust this by specifying -n CPU# and --mem Megabytes. You may request up to 5 hours of CPU time for interactive jobs.
salloc --time 5:00:00 --partition cpu2019
Always use salloc or srun to start an interactive job. Do not SSH directly to a compute node as SSH sessions will be refused without an active job running.
Running non-interactive jobs (batch processing)
Production runs and longer test runs should be submitted as (non-interactive) batch jobs, in which commands to be executed are listed in a script (text file). Batch jobs scripts are submitted using the sbatch command, part of the Slurm job management and scheduling software. #SBATCH directive lines at the beginning of the script are used to specify the resources needed for the job (cores, memory, run time limit and any specialized hardware needed).
Most of the information on the Running Jobs (external link) page on the Compute Canada web site is also relevant for submitting and managing batch jobs and reserving processors for interactive work on ARC. One major difference between running jobs on the ARC and Compute Canada clusters is in selecting the type of hardware that should be used for a job. On ARC, you choose the hardware to use primarily by specifying a partition, as described below.
Selecting a Partition
There are some aspects to consider when selecting a partition including:
- Resource requirements in terms of memory and CPU cores
- Hardware specific requirements, such as GPU or CPU Instruction Set Extensions
- Partition resource limits and potential wait time
- Software support parallel processing using Message Passing Interface (MPI), OpenMP, etc.
- Eg. MPI for parallel processing can distribute memory across multiple nodes, per-node memory requirements could be lower. Whereas, OpenMP or single process code that is restricted to one node would require a higher memory node.
- Note: MPI code running on hardware with Omni-Path networking should be compiled with Omni-Path networking support. This is provided by loading the
openmpi/2.1.3-opaoropenmpi/3.1.2-opamodules prior to compiling.
Since resources that are requested are reserved for your job, please request only as much CPU and memory as your job requires to avoid reducing the cluster efficiency. If you are unsure which partition to use or the specific resource requests that are appropriate for your jobs, please contact us at support@hpc.ucalgary.ca and we would be happy to work with you.
| Partition | Description | Cores/node | Memory Request Limit | Time Limit | GPU | Networking |
|---|---|---|---|---|---|---|
| cpu2019 | General Purpose Compute | 40 | 185,000 MB | 7 days ‡ | 100 Gbit/s Omni-Path | |
| bigmem | Big Memory Compute | 80 | 3,000,000 MB | 24 hours ‡ | 100 Gbit/s Omni-Path | |
| gpu-v100 | GPU Compute | 80 | 753,000 MB | 24 hours ‡ | 2 | 100 Gbit/s Omni-Path |
| apophis† | Private Research Partition | 40 | 185,000 MB | 7 days ‡ | 100 Gbit/s Omni-Path | |
| apophis-bf† | Back-fill Compute | 40 | 185,000 MB | 5 hours ‡ | 100 Gbit/s Omni-Path | |
| razi† | Private Research Partition | 40 | 185,000 MB | 7 days ‡ | 100 Gbit/s Omni-Path | |
| razi-bf† | Back-fill Compute | 40 | 185,000 MB | 5 hours ‡ | 100 Gbit/s Omni-Path | |
| pawson† | Private Research Partition | 40 | 185,000 MB | 7 days ‡ | 100 Gbit/s Omni-Path | |
| pawson-bf† | Back-fill Compute | 40 | 185,000 MB | 5 hours ‡ | 100 Gbit/s Omni-Path | |
| theia† | Private Research Partition | 28 | 188,000 MB | 7 days ‡ | 40 Gbit/s InfiniBand | |
| theia-bf† | Back-fill Compute | 28 | 188,000 MB | 5 hours ‡ | 40 Gbit/s InfiniBand | |
| synergy† | Private Research Partition | 14 | 245,000 MB | 7 days ‡ | 40 Gbit/s InfiniBand | |
| synergy-bf† | Back-fill Compute | 14 | 245,000 MB | 5 hours ‡ | 40 Gbit/s InfiniBand | |
| cpu2013 | Legacy General Purpose Compute | 16 | 120000 | 7 days ‡ | 40 Gbit/s InfiniBand | |
| lattice | Legacy General Purpose Compute | 8 | 12000 | 7 days ‡ | 40 Gbit/s InfiniBand | |
| parallel | Legacy General Purpose Compute | 12 | 23000 | 7 days ‡ | 40 Gbit/s InfiniBand | |
| single | Legacy Single-Node Job Compute | 8 | 12000 | 7 days ‡ | 40 Gbit/s InfiniBand |
Hardware resource and job policy limits
In addition to the hardware limitations, please be aware that there may also be policy limits imposed on your account for each partition. These limits restrict the number of cores, nodes, or GPUs that can be used at any given time. Since the limits are applied on a partition-by-partition basis, using resources in one partition should not affect the available resources you can use in another partition.
These limits can be listed by running:
$ sacctmgr show qos format=Name,MaxWall,MaxTRESPU%20,MaxSubmitJobs
Name MaxWall MaxTRESPU MaxSubmit
---------- ----------- -------------------- ---------
normal 7-00:00:00 2000
breezy 3-00:00:00 cpu=384 2000
gpu 7-00:00:00 13000
cpu2019 7-00:00:00 cpu=240 2000
gpu-v100 1-00:00:00 cpu=80,gres/gpu=4 2000
single 7-00:00:00 cpu=408,node=75 2000
razi 7-00:00:00 2000
Specifying a partition in a job
One you have decided which partitions best suits your computation, you can select one or more partition on a job-by-job basis by including the partition keyword for an SBATCH directive in your batch job. Multiple partitions should be comma separated. If you omit the partition specification, the system will try to assign your job to appropriate hardware based on other aspects of your request.
In some cases, you really should specify the partition explicitly. For example, if you are running single-node jobs with thread-based parallel processing requesting 8 cores you could use:
#SBATCH --mem=0 ❶
#SBATCH --nodes=1 ❷
#SBATCH --ntasks=1 ❸
#SBATCH --cpus-per-task=8 ❹
#SBATCH --partition=single,lattice ❺
A few things to mention in this example:
--mem=0allocates all available memory on the compute node for the job. This effectively allocates the entire node for your job.--nodes=1allocates 1 node for the job--ntasks=1your job has a single task--cpus-per-task=8asks for 8 CPUs per task. This job in total will request 8 * 1, or 8 CPUs.--partition=single,latticespecifies that this job can run on either single or lattice.
Suppose that your job requires at most 8 CPU cores and 10 GB of memory. The above Slurm request would be valid and optimal since your job fits neatly in a single node on the single and parallel partition. However, if you failed to specify the partition, Slurm may try to schedule your job to a partition with larger nodes, such as cpu2019 where each node has 40 cores and 190 GB of memory. If your job is scheduled on such a node, your job will be effectively wasting 32 cores and 180 GB of memory because --mem=0 not only requests for 190 GB on this node, but also prevents other jobs from being scheduled on the same node.
If you don't specify a partition, please give greater thought to the memory specification to make sure that the scheduler will not assign your job more resources than are needed.
Parameters such as --ntasks-per-cpu, --cpus-per-task, --mem and --mem-per-cpu> have to be adjusted according to the capabilities of the hardware also. The product of --ntasks-per-cpu and --cpus-per-task should be less than or equal to the number given in the "Cores/node" column. The --mem> parameter (or the product of --mem-per-cpu and --cpus-per-task) should be less than the "Memory limit" shown. If using whole nodes, you can specify --mem=0 to request the maximum amount of memory per node.
Examples
Here are some examples of specifying the various partitions.
As mentioned in the Hardware section above, the ARC cluster was expanded in January 2019. To select the 40-core general purpose nodes specify:
#SBATCH --partition=cpu2019
To run on the Tesla V100 GPU-enabled nodes, use the gpu-v100 partition. You will also need to include an SBATCH directive in the form --gres=gpu:n to specify the number of GPUs, n, that you need. For example, if the software you are running can make use of both GPUs on a gpu-v100 partition compute node, use:
#SBATCH --partition=gpu-v100 --gres=gpu:2
For very large memory jobs (more than 185000 MB), specify the bigmem partition:
#SBATCH --partition=bigmem
If the more modern computers are too busy or you have a job well-suited to run on the compute nodes described in the legacy hardware section above, choose the cpu2013, Lattice or Parallel compute nodes by specifying the corresponding partition keyword:
#SBATCH --partition=cpu2013 #SBATCH --partition=lattice #SBATCH --partition=parallel
There is an additional partition called single that provides nodes similar to the lattice partition, but, is intended for single-node jobs. Select the single partition with
#SBATCH --partition=single
Time limits
Use the --time directive to tell the job scheduler the maximum time that your job might run. For example:
#SBATCH --time=hh:mm:ss
You can use scontrol show partitions or sinfo to see the current maximum time that a job can run.
$ scontrol show partitions
PartitionName=single
AllowGroups=ALL AllowAccounts=ALL AllowQos=ALL
AllocNodes=ALL Default=NO QoS=single
DefaultTime=NONE DisableRootJobs=NO ExclusiveUser=NO GraceTime=0 Hidden=NO
MaxNodes=UNLIMITED MaxTime=7-00:00:00 MinNodes=1 LLN=NO MaxCPUsPerNode=UNLIMITED
Nodes=cn[001-168]
PriorityJobFactor=1 PriorityTier=1 RootOnly=NO ReqResv=NO OverSubscribe=NO
OverTimeLimit=NONE PreemptMode=OFF
State=UP TotalCPUs=1344 TotalNodes=168 SelectTypeParameters=NONE
DefMemPerNode=UNLIMITED MaxMemPerNode=UNLIMITED
Alternatively, with sinfo under the TIMELIMIT column:
$ sinfo
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
single up 7-00:00:00 1 drain* cn097
single up 7-00:00:00 1 maint cn002
single up 7-00:00:00 4 drain* cn[001,061,133,154]
...
Support
|
Need Help or have other ARC Related Questions? For all general RCS related issues, questions, or comments, please contact us at support@hpc.ucalgary.ca.
|
Please don't hesitate to contact us directly by email if you need help using ARC or require guidance on migrating and running your workflows to ARC.