Bioinformatics applications: Difference between revisions
Jump to navigation
Jump to search
Created page with "== Strategies to write efficient bioinformatics workflows for the ARC high performance computing (HPC) cluster == ===== One of the challenges to deal with big genomics data..." |
No edit summary |
||
| Line 1: | Line 1: | ||
== Strategies to write efficient bioinformatics workflows for the ARC high performance computing (HPC) cluster == | == Strategies to write efficient bioinformatics workflows for the ARC high performance computing (HPC) cluster == | ||
| Line 6: | Line 7: | ||
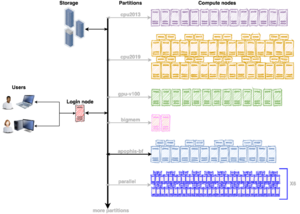
===== The ARC cluster offers a variety of hardware gears to suit the requirements of any workflow. As a first step, review the compute resources on the ARC cluster (weblink). It will facilitate the process of choosing the appropriate partition depending on the nature of the workflow. For example, choose the gpu-v100 partition for GPU accelerators or the bigmem partition for a memory intensive workflow. Each partition has multiple compute nodes with similar hardware specifications. The illustration below shows that gpu-v100 partition has 13 compute nodes, cpu2019 partition has 40 compute nodes, cpu2013 has 14 compute nodes and so on.===== | ===== The ARC cluster offers a variety of hardware gears to suit the requirements of any workflow. As a first step, review the compute resources on the ARC cluster (weblink). It will facilitate the process of choosing the appropriate partition depending on the nature of the workflow. For example, choose the gpu-v100 partition for GPU accelerators or the bigmem partition for a memory intensive workflow. Each partition has multiple compute nodes with similar hardware specifications. The illustration below shows that gpu-v100 partition has 13 compute nodes, cpu2019 partition has 40 compute nodes, cpu2013 has 14 compute nodes and so on.===== | ||
[[File:ARC ClusterV2.png|frameless|center|The ARC cluster is a diverse collection of hardwares ]] | |||
This section will review the SLURM job submission scripts for the bioinformatics applications below. | This section will review the SLURM job submission scripts for the bioinformatics applications below. | ||
Revision as of 05:33, 8 June 2021
Strategies to write efficient bioinformatics workflows for the ARC high performance computing (HPC) cluster
One of the challenges to deal with big genomics data set is their long runtimes. The effective and efficient use of the computing resources on the Advanced Research Computing (ARC) cluster can speed up the runtimes.
The ARC cluster offers a variety of hardware gears to suit the requirements of any workflow. As a first step, review the compute resources on the ARC cluster (weblink). It will facilitate the process of choosing the appropriate partition depending on the nature of the workflow. For example, choose the gpu-v100 partition for GPU accelerators or the bigmem partition for a memory intensive workflow. Each partition has multiple compute nodes with similar hardware specifications. The illustration below shows that gpu-v100 partition has 13 compute nodes, cpu2019 partition has 40 compute nodes, cpu2013 has 14 compute nodes and so on.
This section will review the SLURM job submission scripts for the bioinformatics applications below.
- FastQC - A high throughput sequence QC analysis tool
- Burrows-Wheeler Aligner (BWA)
- Samtools